
MolES faculty member and bioengineering professor Barry Lutz, in partnership with Dr. Matthew Thompson, a UW professor of family medicine and global health, is pioneering at home test kits for the Seattle Coronavirus Assessment Network to respond to the COVID-19 pandemic.
Read more about how the Lutz lab is developing new ways to rapidly test for COVID-19.
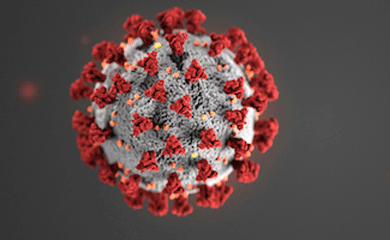
In response to the COVID-19 pandemic, MolES faculty have pivoted their research to address the novel coronavirus, SARS-CoV-2. They are leveraging molecular engineering approaches and tools to develop improved diagnostics, targeted treatment strategies, and a better understanding of the virus. We highlight a few of these projects here.

Using advanced instrumentation in the Molecular Analysis Facility, researchers in the lab of MolES faculty member and materials science & engineering professor Christine Luscombe have discovered that Salish Sea oysters may not contain as many microplastic contaminants as previously thought.

Jason Fontana, a molecular engineering Ph.D. student in the labs of chemical engineering professor James Carothers and chemistry professor Jesse Zalatan, has identified features of bacterial genes that impose strict requirements on CRISPR-Cas transcriptional activation tools. This work defines new strategies to effectively regulate gene expression in bacteria, bringing researchers closer to their goal of using bacteria to produce valuable biosynthetic products. Read this Q&A with Jesse Zalatan featured on the Science in Seattle blog.

A team was led by Dr. Alshakim Nelson, an assistant professor of chemistry at the UW, and Dr. Hal Alper, a professor of chemical engineering at the University of Texas, developed a new method that combines the bioactivity of microbes and a 3D-printed, synthetic hydrogel "” a water-based gel structure "” to create desired chemical compounds. The products can vary from pharmaceuticals to nutraceuticals, alluding to the vast potential for this new finding.
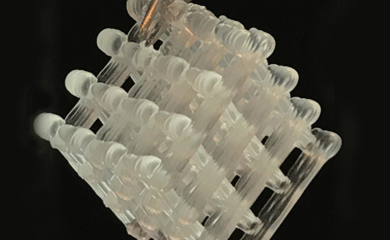
Researchers in the lab of MolES faculty member and professor of chemistry Al Nelson along with collaborators at the University of Texas unveiled a new way to produce medicines and chemicals and preserve them using portable "biofactories" that are embedded in water-based gels known as hydrogels. The approach could help people in remote villages or on military missions, where the absence of pharmacies, doctor's offices or even basic refrigeration makes it hard to access critical medicines and other small-molecule compounds.
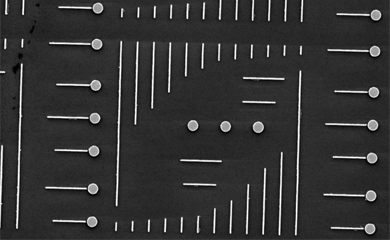
A team led by MolES faculty member David Masiello and scientists from the University of Notre Dame used recent advances in electron microscopy to observe Fano interferences "” a form of quantum-mechanical interference by electrons "” directly in a pair of metallic nanoparticles.
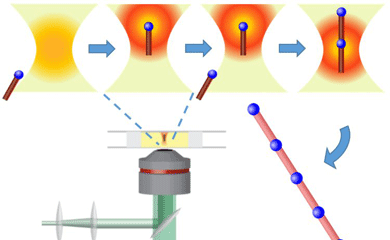
A team led by MolES faculty member Peter Pauzauskie, a professor of materials science and engineering, has developed a method that could make reproducible manufacturing at the nanoscale possible. The team adapted a light-based technology employed widely in biology "” known as optical traps or optical tweezers "” to operate in a water-free liquid environment of carbon-rich organic solvents, thereby enabling new potential applications.
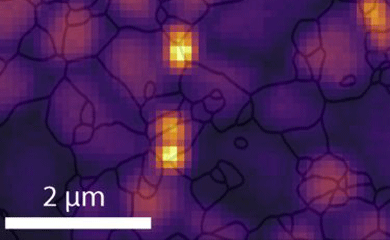
A team led by David Ginger, professor of chemistry and MolES faculty member, has developed a way to map strain in lead halide perovskite solar cells. Their approach shows that misorientation between microscopic perovskite crystals is the primary contributor to the buildup of strain within the solar cell, which creates small-scale defects in the grain structure, interrupts the transport of electrons within the solar cell, and ultimately leads to heat loss through a process known as non-radiative recombination.
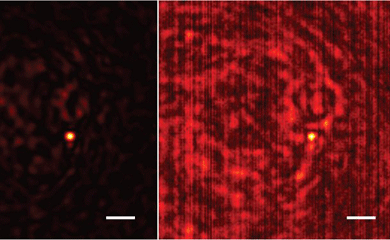
A team led by MolES faculty member Arka Majumdar, an assistant professor of electrical and computer engineering and physics, has designed and tested a 3D-printed metamaterial that can manipulate light with nanoscale precision. As they report in a paper published October 4 in the journal Science Advances, their designed optical element focuses light to discrete points in a 3D helical pattern.